 |


![]()
Proteins are inherently dynamic, and this property is critical for the allostery they exhibit. This means that it is possible to use small molecules to both activate and inhibit protein function at a distance from the active site. We are using an integrative chemical biology approach to characterize the allosteric circuitry of inflammatory and apoptotic caspases.
One goal is to develop allosteric activators of the apoptotic caspases (-3, -6, -7, -8 and -9) so that we may trigger each selectively and understand their roles in apoptosis and other non-apoptotic functions such as stem cell differentiation. Not only will these compounds be important as tools, but also as potential drug leads for cancer treatment.
Active sites of enzymes frequently pose problems in yielding drug-like chemotypes, and often the specificity profiles of homologs overlap. Such is the case for the apoptotic and inflammatory caspases. There are no selective compounds available which allow us to dissect apart their roles in driving pro-inflammatory and pro-apoptotic protein processing. For example, we are interested in developing allosteric inhibitors for the inflammatory caspases (-1, and -5) and the apoptotic caspases (-3 and -7) in order to better understand their importance in driving innate immune and apoptotic functions.
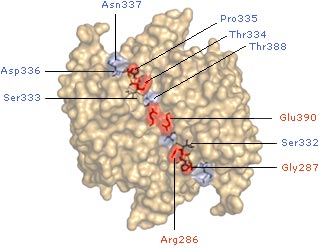
We have employed a powerful site-directed screening method called disulfide-trapping, or tethering, that has allowed us to identify specific inhibitors for caspase-1 that bind to a cysteine in a cavity about 15 Å from the active site. Structural studies show that the compounds trap a natural “off” state of matured caspase-1. These allosteric inhibitors are reversible and highly selective for caspase-1 over caspases–5. Mutational and kinetic analysis show that there is a critical hydrogen-bonding circuit that radiates from this allosteric site to the active sites and that the protease exhibits positive cooperativity that is unique among proteases.
We hypothesize that this allosteric transition and cavity are commonly held in the caspase family. We are now using these compounds to trap specific allosteric transitions in caspases that can be visualized by x-ray crystallography and studied by mutational methods. This will enable identification of common features that control the allosteric circuitry in this highly regulated enzyme family. These studies are providing better validation of these enzymes as potential drug targets for inflammatory diseases, and fully characterizing a novel allosteric site that may be more suitable than the active site for drug discovery efforts.
Wells Lab © 2009 | Site Map