
2018
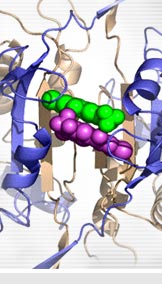
Keedy D.A., Hill Z.B., Biel J.T., Kang E., Rettenmaier T.J., Brandao-Neto J., Pearce N.M., von Delft F., Wells J.A., Fraser J.S. (2018)
"An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering."
eLife. in press
![]()
Pollock S.B., Hu A., Mou Y.M., Martinko A.J., Julien O., Hornsby M., Ploder L, Adams J.J., Geng H., Muschen M., Sidhu S., Moffat J, Wells J.A. (2018)
"Highly multiplexed and quantitative cell surface protein profiling using genetically barcoded antibodies."
Proc. Natl. Acad. Sci. U.S.A. 115:2836-2841
![]()
Kroemer G., et al., Wells J.A., Wood W., Yuan J., Zakeri Z., Zhivotovsky B., Zitvogel L., Melino G. and Kroemer G. (2018)
"Molecular mechanisms of cell death: recommendations of the Nomenclature Committee on Cell Death 2018."
Cell Death Differ. 25:486-541
![]()
Martinko A.J., Truillet C., Julien O., Diaz J.E., Horlbeck, M.A., Whiteley G., Blonder J., Weissman J.S., Bandyopadhyay S., Evans M.J., Wells J.A. (2018)
"Targeting RAS Driven Human Cancer Cells with Antibodies to Upregulated and Essential Cell-Surface Proteins."
eLife. 115:2836-2841
![]()
eLife Press Release
2017
v, Martinko A.J., Nguyen D.P., Wells J.A. (2017)
"Human Antibody-Based Chemically Induced Dimerizers for Cell Therapeutic Applications."
Nat. Chem. Biol. 2:112-117
![]()
F1000 recommendation
Weeks A.M. and Wells J.A. (2017)
"Engineering peptide ligase specificity by proteomic identification of ligation sites."
Nat. Chem. Biol. 14:50-57
![]()
Nat. Chem. Biol. News and Views
F1000 recommendation
Srinivasan S.R., Cesa L.C., Li X., Julien O., Zhuang M., Shao H., Chung J., Maillard I., Wells J.A., Duckett C., Gestwicki J.E. (2017)
"Heat Shock Protein 70 (Hsp70) Suppresses RIP1-dependent Apoptotic and Necroptotic Cascades."
Mol. Cancer Res. 17-0408
![]()
Smart A.D., Pache R.A., Thomsen N., Kortemme T., Davis G.W., Wells J.A. (2017)
"Engineering a Light-Activated Caspase-3 for Precise Ablation of Neurons in Vivo."
Proc. Natl. Acad. Sci. U.S.A. 114:E8174-E8183
![]()
ACS Chem. Bio. Spotlight
Diaz J.E., Morgan C.W., Minogue C.E., Hebert A.S., Coon J.J., Wells J.A. (2017)
"A Split-Abl Kinase for Direct Activation in Cells."
Cell Chem. Bio. 24:1-9
![]()
Cell Chem. Bio. Preview
Liu T.Y., Huang H.H., Wheeler D., Xu Y., Wells J.A., Song Y.S., Wiita A.P. (2017)
"Time-Resolved Proteomics Extends Ribosome Profiling-Based Measurements of Protein Synthesis Dynamics."
Cell Syst. 4:636-644
![]()
Julien O. and Wells J.A. (2017)
"Caspases and their substrates"
Cell Death Differ. 24:1380-1389
![]()
Lin S., Yang X., Jia S., Weeks A.M., Hornsby M., Lee P.S., Nichiporuk R.V., Iavarone A.T., Wells J.A., Toste F.D., Chang C.J. (2017)
"Redox-based reagents for chemoselective methionine bioconjugation."
Science 355:597-602
![]()
Tran H.L., Lexa K.W., Julien O., Young T.S., Walsh C.T., Jacobson M.P., and Wells J.A. (2017)
"SAR and molecular mechanics reveal the importance of ring entropy
in the biosynthesis and activity of a natural product"
J. Am. Chem. Soc. 139:2541-2544
![]()
Calvo S.E., Julien O., Clauser K.R., Shen H., Kamer K.J., Wells J.A. Mootha V.K. (2017)
"Comparative analysis of mitochondrial N-termini from mouse, human, and yeast"
Mol. Cell. Proteomics. 16:512-523
![]()
2016
Spangler B., Fontaine S.D., Shi Y., Sambucetti L.C., Mattis A.N., Hann B., Wells J.A., and Renslo A.R. (2016)
"A Novel Tumor-Activated Prodrug Strategy Targeting Ferrous Iron is Effective in Multiple Preclinical Cancer Models"
J. Med. Chem 59:11161-11170.
![]()
Ye X., Chan K.C., Waters A.M., Bess M., Harned A., Wei B.R., Loncarek J., Luke B.T., Orsburn B.C., Hollinger B.D., Stephens R.M., Bagni R., Martinko A., Wells J.A., Nissley D.V., McCormick F., Whiteley G. and Blonder J. (2016)
"Comparative proteomics of a model MCF10A-KRasG12V cell line reveals a distinct molecular signature of the KRasG12V cell surface"
Oncotargets 7:86948-86971
![]()
Henager SH., Chu N., Chen Z., Bolduc D., Dempsey D.R., Hwang Y., Wells J.A., Cole P.A. (2016)
"Enzyme-catalyzed expressed protein ligation."
Nat. Methods 13:925-927
![]()
Hill Z.B., Pollock S.B., Zhuang Z., Wells J.A. (2016)
"Direct proximity tagging of small molecule protein targets using an engineered NEDD8 ligase"
J. Am. Chem. Soc. 138:13123-13126
![]()
Wang W., Nguyen L.T., Burlak C., Chegini F., Guo F., Chataway T., Ju S., Fisher O., Miller D.W., Datta D., Wu F., Wu C., Landeru A., Wells J.A., Cookson M.R., Boxer M.B., Thomas C.J, Gai W.P., Ringe D., Petsko G.A., and Hoang Q.Q. (2016)
"Caspase-1 causes truncation and aggregation of the Parkinson disease-associated protein α-synuclein."
Proc. Natl. Acad. Sci. U.S.A. 113:9587-9592
![]()
Spangler B., Morgan C.W., Vander Wal M.N., Chang C.J., Hann B., Wells J.A., and Renslo A.R. (2016)
"A reactivity-based probe of the intracellular labile ferrous iron pool."
Nat. Chem. Biol. 12:680-685
![]()
Seaman J.E., Julien O., Lee P.S., Rettenmaier T.J., Thomsen N.D., Wells J.A. (2016)
"Cacidases: caspases can cleave after aspartate, glutamate and phosphoserine residues"
Cell Death Differ. 23:1717-1726
![]()
F1000 recommendation
Nguyen D.P., Miyaoka Y., Gilbert L.A., Mayerl S.J., Lee B.H., Weissman J.S., Conklin B.R., and Wells J.A. (2016)
"Ligand-binding domains of nuclear receptors facilitate tight control of split CRISPR activity."
Nat. Commun. 7:12009.
![]()
Hill, M.E., MacPherson D.J., Wu P. Julien O., Wells J.A., and Hardy J.A. (2016)
"Reprogramming Caspase‑7 Specificity by Regio-Specific Mutations and Selection Provides Alternate Solutions for Substrate Recognition."
ACS Chem. Biol. 11:1603-1612
![]()
Julien O., Zhuang M., Wiita A.P., O'Donoghue A.J., Knudsen G.M., Craik C.S., and Wells J.A. (2016)
"Quantitative MS-based enzymology of caspases reveals distinct protein substrate specificities, hierarchies, and cellular roles."
Proc. Natl. Acad. Sci. U.S.A. 113:E2001-2010
![]()
Cimermancic P., Weinkam P., Rettenmaier T.J., Bichmann L., Keedy D.A., Woldeyes R.A., Schneidmann D., Demerdash O.N., Mitchell J.C., Wells J.A., Fraser J.S., Sali A. (2016)
"CryptoSite: Expanding the Druggable Proteome by Characterization and Prediction of Cryptic Binding Sites."
J. Mol. Biol. 428:709-719
![]()
2015
Rettenmaier J.T., Fan H., Karpiak J., Doak A., Sali A., Shoichet B., and Wells J.A. (2015)
"Allosteric small-molecule modulators of the protein kinase PDK1 discovered by virtual screening"
J. Med. Chem. 58:8285-8291
![]()
Hornsby M., Paduch M., Miersch S., Sääf A., Matsuguchi T., Lee B., Wypisniak K., Doak A., King D., Usatyuk S., Perry K., Lu V., Thomas W., Luke J., Goodman J., Hoey R.J., Lai D., Griffin C., Li Z., Vizeacoumar F.J., Dong D., Campbell E., Anderson S., Zhong N., Gräslund S., Koide S., Moffat J., Sidhu S., Kossiakoff A., and Wells J.A. (2015)
"A High Through-put Platform for Recombinant Antibodies to Folded Proteins."
Mol. Cell. Proteomics. 10:2833-2847
![]()
Morgan C.W., Diaz J.E., Zeitlin S.G., Gray D.C., and Wells J.A. (2014)
"Engineered cellular gene-replacement platform for selective and inducible proteolytic profiling."
Proc. Natl. Acad. Sci. U.S.A. 112:8344-8349
![]()
Miersch S., Li Z., Hanna R., McLaughlin M.E., Hornsby M., Matsuguchi T., Paduch M., Sääf A., Wells J.A., Koide S., Kossiakoff A., and Sidhu S.S. (2015)
"Scalable high throughput selection from phage-displayed synthetic antibody libraries."
J Vis Exp. 95:51492
![]()
Koerber J.T., Hornsby M.J., and Wells J.A. (2015)
"An Improved Single-chain Fab Platform for Efficient Display and Recombinant Expression."
J. Mol. Biol. 427:576-586
![]()
2014
Rettenmaier T.J., Sadowsky J.D., Thomsen N.D., Chen S.C., Doak A.K., Arkin M.R., and Wells J.A. (2014)
"A small-molecule mimic of a peptide docking motif inhibits the protein kinase PDK1."
Proc. Natl. Acad. Sci. U.S.A. 111:18590-18595
![]()
Ordureau A., Sarraf S.A., Duda D.M., Heo J.M., Jedrykowski M.P., Sviderskiy V.O., Olszewski J.L., Koerber J.T., Xie T., Beausoleil S.A., Wells J.A., Gygi S.P., Schulman B.A., Harper J.W. (2014)
"Quantitative Proteomics Reveal a Feedforward Mechanism for Mitochondrial PARKIN Translocation and Ubiquitin Chain Synthesis."
Mol. Cell. 56:360-375
![]()
Julien O., Kampmann M., Bassik M.C., Zorn J.A., Venditto V.J., Shimbo K., Agard N.J., Shimada K., Rheingold A.L., Stockwell B.R., Weissman J.S., and Wells J.A. (2014)
"Unraveling the mechanism of cell death induced by chemical fibrils."
Nat. Chem. Biol. 10:969-976
![]()
Arkin M.R., Tang Y., Wells J.A. (2014)
"Small-Molecule Inhibitors of Protein-Protein Interactions: Progressing toward the Reality."
Chem. Biol. 21:1102-1114.
![]()
Wiita A.P., Seaman J.E., Wells J.A. (2014)
"Global analysis of cellular proteolysis by selective enzymatic labeling of protein N-termini."
Methods Enzymol. 2014;544:327-358
![]()
Morgan C.W., Julien O., Unger E.K., Shah N.M., Wells J.A. (2014)
"Turning on caspases with genetics and small molecules."
Methods Enzymol. 2014;544:179-213
![]()
Wells J.A. and Kossiakoff A.A. (2014)
"Cell biology. New tricks for an old dimer."
Science. 2014 344:703-704
![]()
Wiita A.P., Hsu G.W., Lu C.M., Esensten J.H., Wells J.A. (2014)
"Circulating Proteolytic Signatures of Chemotherapy-induced Cell Death in Humans Discovered by N-terminal Labeling."
Proc. Natl. Acad. Sci. 111:7594-7599
![]()
Lodge J.M, Rettenmaier J.T., Wells J.A., Pomerantz W.C., and Mapp A.K. (2014)
"FP tethering: a screening technique to rapidly identify compounds that disrupt protein–protein interactions"
Med. Chem. Commun. 5:370-375
![]()
2013
Ostrem J.M., Peters U., Sos M.L., Wells J.A., Shokat K.M. (2013)
"K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions."
Nature. 503:548-551
![]()
Wiita A.P., Ziv E., Wiita P.J., Urisman A., Julien O., Burlingame A.L., Weissman J.S., and Wells J.A. (2013)
"Global cellular response to chemotherapy-induced apoptosis."
eLife. 2:e01236
![]()
Koerber J.T, Thomsen N.D., Hannigan B.T., Degrado W.F., Wells J.A. (2013)
"Nature-inspired design of motif-specific antibody scaffolds."
Nat. Biotechnol. 31:916-921
![]()
Carter P.J., Hazuda D., Wells J.A. (2013)
"Next generation therapeutics."
Curr Opin Chem Biol. 17. 317-319
![]()
Yang C.F., Chiang M.C., Gray D.C., Prabhakaran M., Alvarado M., Juntti S.A., Unger E.K., Wells J.A., Shah N.M. (2013)
"Sexually dimorphic neurons in the ventromedial hypothalamus govern mating in both sexes and aggression in males."
Cell. 153:896-909
![]()
Thomsen N.D., Koerber J.T., Wells J.A. (2013)
"Structural snapshots reveal distinct mechanisms of procaspase-3 and -7 activation."
Proc. Natl. Acad. Sci. 110. 8477-8482
![]()
Datta D., McClendon C.L., Jacobson M.P., Wells J.A. (2013)
"Substrate and inhibitor-induced dimerization and cooperativity in caspase-1 but not caspase-3."
J. Biol. Chem. 288. 9971-9981
![]()
Wang N., Majmudar C.Y., Pomerantz W.C., Gagnon J.K., Sadowsky J.D., Meagher J.L., Johnson T.K., Stuckey J.A., Brooks C.L. 3rd, Wells J.A., Mapp A.K. (2013)
"Ordering a Dynamic Protein Via a Small-Molecule Stabilizer."
J. Am. Chem. Soc. 135, 3363-2266
![]()
Crawford E.D., Seaman J.E., Agard N., Hsu G.W., Julien O., Mahrus S., Nguyen H., Shimbo K., Yoshihara H., Zhuang M., Chalkley R.J., Wells J.A. (2013)
"The DegraBase: a database of proteolysis in healthy and apoptotic human cells."
Mol. Cell. Proteomics 12, 813-824
![]()
Zhuang M., Guan S., Wang H., Burlingame A.L., Wells J.A. (2013)
"Substrates of IAP Ubiquitin Ligases Identified with a Designed Orthogonal E3 Ligase, the NEDDylator"
Mol. Cell. 49, 273-282.
![]()
2012
Crawford E.D., Seaman J.E., Barber A.E., David D.C., Babbitt P.C., Burlingame A.L., Wells J.A. (2012)
"Conservation of caspase substrates across metazoans suggests hierarchical importance of signaling pathways over specific targets and cleavage site motifs in apoptosis"
Cell Death Differ. 19, 2040-2048.
![]()
Zorn J.A., Wolan D.W., Agard N.J., Wells J.A. (2012)
"Fibrils Colocalize Caspase-3 with Procaspase-3 to Foster Maturation"
J. Biol. Chem. 287, 33781-22795.
![]()
Shimbo K., Hsu G.W., Nguyen H., Mahrus S., Trinidad J.C., Burlingame A.L., and Wells J.A. (2012)
"Quantitative profiling of caspase-cleaved substrates reveals different drug-induced and cell-type patterns in apoptosis"
Proc Natl Acad Sci U S A. 109, 12432-12437.
![]()
Agard N.J., Mahrus S., Trinidad J.C., Lynn A., Burlingame A.L., Wells J.A. (2012)
"Global kinetic analysis of proteolysis via quantitative targeted proteomics"
Proc Natl Acad Sci U S A. 109, 1913-1918.
![]()
2011
Zorn J.A., Wille H., Wolan D.W., Wells J.A. (2011)
"Self-Assembling Small Molecules Form Nanofibrils That Bind Procaspase-3 to Promote Activation"
J Am Chem Soc. 133, 19630-19633.
![]()
Gao J. & Wells J.A. (2011)
"Identification of Specific Tethered Inhibitors for Caspase-5"
Chem Biol Drug Des. 79, 209-215.
![]()
Tang Y., Wells J.A. & Arkin M.R. (2011)
"Structural and enzymatic insights into caspase-2 substrate recognition and catalysis"
J Biol Chem. 286, 34147-34154.
![]()
Crawford E.D. & Wells J.A. (2011)
"Caspase substrates and cellular remodeling"
Annu Rev Biochem. 80, 1055-87.
![]()
Sadowsky, J., Burlingame, M., Wolan, D., McClendon, C., Jacobson, M., & Wells, J.A. (2011)
"Turning a protein kinase (PDK1) on or off from a single allosteric site via disulfide trapping"
Proc. Natl. Acad. Sci. USA. 108, 6056-6061.
![]()
Perez-Paya, E., Orzaez, M, Mondragon, L, Wolan, D, Wells, JA, Messeguer, A, Vincent, MJ. (2011)
"Molecules that modulate Apaf-1 activity"
Med Res Rev. 31, 649-675.
![]()
2010
Gray, D.C., Mahrus, S. & Wells. J.A. (2010)
"Activation of specific apoptotic caspases with an engineered small molecule activated protease"
Cell 142, 637-646.
![]()
Barkan, D.T., Hostetter, D.R., Mahrus, S., Pieper, U., Wells, J.A., Craik, C.S. & Sali, A. (2010)
"Prediction of protease substrates using sequence and structure features"
Bioinformatics 26, 1714-22.
![]()
Agard, N.J, Maltby, D. & Wells, J.A. (2010)
"Inflammatory stimuli regulate caspase substrate profiles"
Mol. Cell. Proteomics 5, 880-893.
![]()
Zorn, JA, & Wells, JA. (2010)
"Turning enzymes ON with small molecules"
Nat Chem Biol 3, 179-188.
![]()
Wildes, D & Wells, J.A. (2010)
"Sampling the N-terminal proteome of human blood"
Proc. Natl. Acad. Sci. USA 107, 4561-4566.
![]()
2009
Wolan DW, Zorn JA, Gray DC, Wells JA. (2009)
"Small-molecule activators of a proenzyme"
Science 326, 853-858.
![]()
Agard, N. & Wells, J.A. (2009)
"Methods for the proteomic identification of protease substrates"
Curr Opin Chem Biol 13, 503-509.
![]()
Hardy, J. & Wells, J.A. (2009)
"Dissecting an allosteric switch in caspase-7 using chemical and mutational probes"
Journal of Biological Chemistry 284(38), 26063-26069.
Journal of Biological Chemistry 284(38), 26063-26069.
![]()
Gao J, Sidhu SS, Wells JA. (2009)
"Two-state selection of conformation-specific antibodies"
Proc. Natl. Acad. Sci USA 106, 3071-3076.
![]()
2008
Yoshihara HA, Mahrus S, Wells JA. (2008) Bioorg Med Chem Lett. 18, 6000-6003. "Tags for labeling protein N-termini with subtiligase for proteomics"
Mahrus S, Trinidad JC, Barkan DT, Sali A, Burlingame AL, Wells JA. (2008)
"Global sequencing of proteolytic cleavage sites in apoptosis by specific labeling of protein N termini"
Cell 134, 866-876.
![]()
Datta D, Scheer JM, Romanowski MJ, Wells JA. (2008) J Mol Biol. 381,
1157-1167. "An allosteric circuit in caspase-1"
![]()
2007
Wells JA, McClendon CL. (2007) Nature 450, 1001-1009. "Reaching
for high-hanging fruit in drug discovery at protein-protein interfaces"
![]()
2006
Thanos, C.D., DeLano, W.L. and Wells, J.A. (2006)
"Hot-spot mimicry of a cytokine receptor by a small molecule"
![]()
Scheer, J., Romanowski, M. and Wells, J.A. (2006) Proc. Natl. Acad. Sci. USA 103, 7595-7600.
"A Common Allosteric Site and Mechanism in Caspases"
![]()
2005
Scheer, J., Wells, J.A. and Romanowski, M. (2005) Protein Expr. Purif. 148-153.
"Malonate-assisted purification of human caspases"
![]()
Buck, E. and Wells, J.A. (2005) Proc. Natl. Acad. Sci. USA 102, 2719-2724.
"Disulfide Trapping to Localize Small Molecule Agonists and Antagonists for the Complement 5a Receptor"
![]()
Buck, E., Bourne, H. and Wells, J.A. (2005) J. Biol. Chem 280, 4009-4012.
"Site-specific Disulfide Capture of Agonist and Antagonist Peptides to the C5a Receptor"
![]()
2004
Hardy, J.A. and Wells, J.A. (2004) Curr Opin Struct Biol., 706-715.
"Searching for new allosteric sites in enzymes."
![]()
Hardy, J.A., Lam, J., Nguyen, J.T., O’Brien, T and Wells, J.A. (2004)
"Discovery of an Allosteric Site in Caspases". Proc. Natl. Acad. Sci. USA 101, 12461-12466.
![]()
Erlanson, D.A., Wells, J.A. and Braisted, A.B. (2004)
"TETHERING: Fragment-based Drug Discovery". Annu. Rev. Biophys. Biomol Struct. 33, 199-233.
![]()
Stroud, R.M. and Wells, J.A. (2004) Science STKE, 2004, re7.
"Mechanistic Diversity of Cytokine Signaling Across Cell Membranes"
![]()
Arkin, M.R. and Wells, J.A. (2004) Nature Rev. Drug Disc. 3, 301-317.
"Small Molecule Inhibitors of Protein-Protein Interactions: Progressing Towards the Dream"
![]()
2003
Thanos, C.D., Randal, M., and Wells, J.A. (2003) J. Am. Chem. Soc., 125, 15280-15281.
"Potent Small-molecule Binding to a Dynamic Hot-spot on IL-2"
![]()
Nguyen, J. and Wells, J.A. (2003)
Machinery as a Mechanism to target Cancer Cells"
Proc. Natl. Acad. Sci USA, 100, 7533-7538. "Direct Activation of the Apoptosis
![]()
Arkin, M.R., Randal, M., DeLano, W., Hyde, J., Loung, T.N., Oslob, J.D., Raphael, D.R., Taylor, L., Wang, J., McDowell, R.S., Wells, J.A., Braisted, A.C. (2003)
"Binding of Small Molecules to an Adaptive Protein:Protein Interface"
Proc. Natl. Acad. Sci USA 100, 1603-1608.
![]()
Selected Earlier Publications
Erlanson, D.A., Braisted, A.C., Raphael, D.R., Randal, M., Stroud, R.M., Gordon, E.M. and Wells, J.A. (2000) Proc. Natl. Acad. Sci. USA 97, 9367-9372. "Site-Directed Ligand Discovery"
DeLano, W.L., Ultsch, M.H., De Vos, A.M. and Wells, J.A. (2000) Science 287, 1279-1283. "Convergent Solutions to Binding at a Protein-Protein Interface"
Atwell, S. and Wells, J.A. (1999) Proc. Natl. Acad. Sci USA 96, 9497-9502. "Selection for Improved Subtiligases by Phage Display"
Ballinger, M.D. and Wells, J.A. (1998) Nature Struct. Biol. 5, 938-940. "Will Any Dimer Do?" News and Views
Clackson, T., Ultsch, M., Wells, J.A. and De Vos, A.A. (1998) J. Mol. Biol. 277, 1111-1128. "Structural and Functional Analysis of the 1:1 Growth Hormone:Receptor Complex Reveals the Molecular Basis for Receptor Affinity"
Wiesmann, C., Fuh, G., Christinger, H.W., Eigenbrot, C., Wells, J.A., De Vos, A.M. (1997) Cell 91, 695-704. "Crystal Structure at 1.7Å resolution of VEGF in Complex with Domain 2 of the Flt-1 Receptor"
Atwell, S., Ultsch, M., De Vos, A.M. and Wells, J.A. (1997) Science 278, 1124-1127 "Structural Plasticity in a Remodeled Protein-Protein Interface"
Ellman, J., Stoddard, B., and Wells, J.A. (1997) Proc. Natl. Acad. Sci. USA 94, 2779-2782. "Combinatorial Thinking in Chemistry and Biology"
Wells, J.A. (1996) Science 273, 449-450. "Hormone Mimicry". Review.
Li, B., Tom, J.Y.K., Oare, D., Yen, R., Fairbrother, W., Wells, J.A., and Cunningham, B.C. (1995) Science 270, 1657-1660. "Minimization of a Polypeptide Hormone"
Wells, J.A. (1995) Biotechnology 13, 647-651. "Structural and Functional Epitopes in the Growth Hormone Receptor Complex"
Clackson, T. and Wells, J.A. (1995) Science 267, 383-386. " A Hot Spot of Binding Energy in a Hormone-Receptor Interface"
Chang, T.K., Jackson, D.Y., Burnier, J.P. and Wells, J.A. (1994) Proc. Natl. Acad. Sci. USA 91, 12544-12548. "Subtiligase: a Tool for Semi-synthesis of Proteins"
Jackson, D.Y., Burnier, J., Quan, C., Stanley, M., Tom, J. and Wells, J.A. (1994) Science 266, 243-247. "A Designed Peptide Ligase for Total Synthesis of Ribonuclease A with Unnatural Catalytic Residues."
Matthews, D.J. and Wells, J.A. (1993) Science 260, 1113-1117. "Substrate Phage: Selection of Protease Substrates by Monovalent Phage Display."
Fuh, G. Cunningham, B.C., Fukunaga, R., Nagata, S., Goeddel, D.V., and Wells, J.A. (1992) Science, 256, 1677-1680. "Rational Design of Potent Antagonists to the Human Growth Hormone Receptor."
Cunningham, B.C., Ultsch, M., DeVos, A.M., Mulkerrin, M.G., Clauser, K.R., Wells, J.A. (1991) Science 254, 821-825. "Dimerization of the Extracellular Domain of the Human Growth Hormone Receptor by a Single Hormone Molecule."
Lowman, H.B., Bass, S., Simpson, N., Wells, J.A. (1991) Biochemistry 30, 10832-10838 "Engineering a Hormone for Improved Receptor binding Affinity by Monovalent Phage Display."
Cunningham, B.C., Mulkerrin, M.G., and Wells, J.A. (1991) Science 253, 545-548, "Dimerization of Human Growth Hormone by Zinc."
Cunningham, B.C., and Wells, J.A. (1991) Proc. Natl. Acad. Sci. USA 88, 3407-3411. "Rational design of receptor specific variants of human growth hormone."
Cunningham, B.C., Bass, S., Fuh, G., and Wells, J.A. (1990) Science 250, 1709-1712 "Zinc mediation of the binding of human growth hormone to the human prolactin receptor."
Cunningham, B.C., Henner, D.J. and Wells, J.A. (1990) Science 247, 1461-1465. "Engineering human prolactin to bind to the human growth hormone receptor."
Cunningham, B.C. and Wells, J.A., (1989) Science 244, 1081-1085. "High-resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis."
Cunningham, B.C., Jhurani, P., Ng, P. and Wells, J.A., (1989) Science 243, 1330-1336. "Receptor and antibody epitopes in human growth hormone identified by homolog-scanning mutagenesis."
Mitchinson, C. and Wells, J.A. (1989) Biochemistry 28, 4807-4815. "Protein engineering of disulfide bonds in subtilisin."
Wells, J.A. and Estell, D.A. (1988) Trends in Biochem. Sci. 13, 291-297. "Subtilisin - an enzyme designed to be engineered."
Carter, P. and Wells, J.A. (1988) Nature 332, 564-568. "Dissecting the catalytic triad of a serine protease."
Carter, P., and Wells, J.A. (1987) Science 24, 394-399. "Engineering substrate specificity by substrate-assisted catalysis."
Wells, J.A., Cunningham, B.C., Graycar, T.P., and Estell, D.A. (1987). Proc. Natl. Acad. Sci. USA 84, 5167-5171. "Recruitment of substrate specificity properties from one enzyme into a related one by protein engineering."
Wells, J.A., Powers, D.B., Bott, R.R., Graycar, T.P., and Estell, D.A. (1987) Proc. Natl. Acad. Sci. USA 84, 1219-1223. "Designing substrate specificity by protein engineering of electrostatic interactions."
Estell, D.A., Graycar, T.P., Miller, J.V., Powers, D.B., Burnier, J.P., Ng, P.G., and Wells, J.A. (1986) Science 233, 659-663. "Probing steric and hydrophobic effects on enzyme-substrate interactions by protein engineering."
Wells, J.A., Powers, D.B., Bott, R.R., Katz, B.A., Ultsch, M.H., Kossiakoff, A.A., Power, S.D., Adams, R.M., Heyneker, H.H., Cunningham, B.C., Miller, J.V., Graycar, T.P., and Estell, D.A. (1987) in Protein Engineering (A.R. Liss) in press, "Protein engineering of subtilisin."
Wells, J.A. and Powers, D.B. (1986) J. Biol. Chem. 261, 6564-6570. "In vivo formation and stability of engineered disulfide bonds in subtilisin."
Wells, J.A. and Yount, R.G. (1979) Proc. Natl. Acad. Sci. USA 76, 4966-4970. "Active site trapping of nucleotides by crosslinking two sulfhydryls in myosin sub-fragment 1."